Antibody drugs are among the most promising therapies and experienced explosive growth as new drugs have been approved for various diseases. Recent advances in antibody discovery technologies have paved way for development of a plethora of new antibody therapeutics.
In the past antibodies are discovered by well-established in vivo immunization procedures or in vitro antibody library screening approaches. However, the exacerbate cost and relatively high failure rate of the traditional path of drug discovery and development has prompted the urgent need for in silico approaches, especially structure-guided approach. The expansion of data repositories including protein structures, sequences and NGS has significantly increased the availability of large-scale data for antibody discovery. The availability of such data sets can provide a significant impact in streamlining antibody discovery process.
Synbio Technologies provides fast, flexible and customized one-stop solutions for antibody discovery. GenoAb platform includes hybridoma/monoclonal antibody sequencing, immune repertoire sequencing and phage display. SynoAb platform is a structure-based in silico antibody discovery method. The platform can screen early hits for antigens for wet-lab evaluation, followed by affinity maturations using controlled permutation libraries. The synergy from GenoAb and SynoAb will offer highly flexible supports and services for complicated antibody discovery.
Featured Advantages
1. Powerful drug-like antibody bio-computation
- Cloud computing
- Supercomputing
2. Proprietary software platforms
- High-throughput antibody modeling
- Effective antigen-antibody docking
3. Facilitate the development of “biologically superior” antibody drugs
- Antibodies humanization
- Affinity maturation
- Full human antibody library design
4. Therapeutic antibodies have been licensed
Service Specifications
| Service | Details | Turnaround Time | Deliverables | Price |
|---|---|---|---|---|
| Syno® Ab Antibody Design and Production |
|
2-3 Weeks |
|
Inquire |
* Customers can provide the antigen name or gene ID directly; If you have antigen crystal structure, please contact us directly.
Service Flow

Case Study
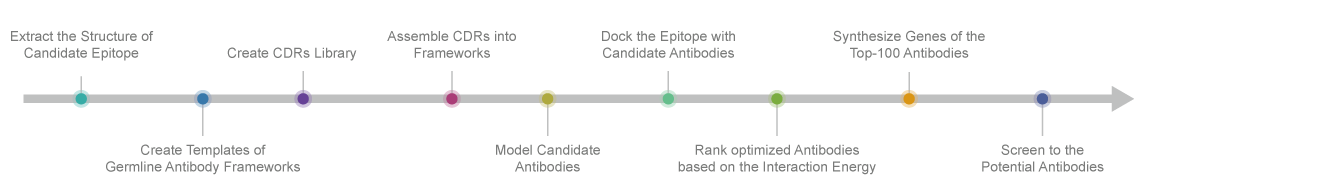
1. Extract potential epitopes based on the interface between target and ligand protein.
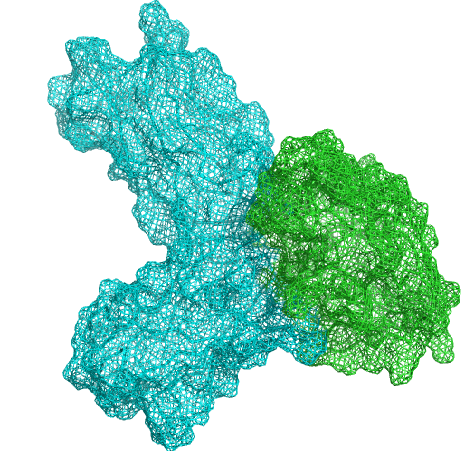
2. Construct antibody library based on 3D structure.
3. Place antibodies based on surface complementary with target.
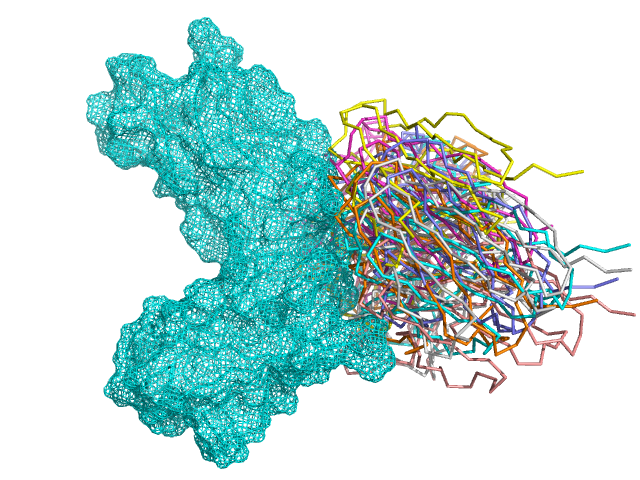
4. Refine “hitting” antibody- target to increase its stability according to amino acids bias at different locations in antibody.
5. Select the best antibody hits according to evaluation, optimize DNA and then synthesize gene/library using Syno® 3.0 synthesis.
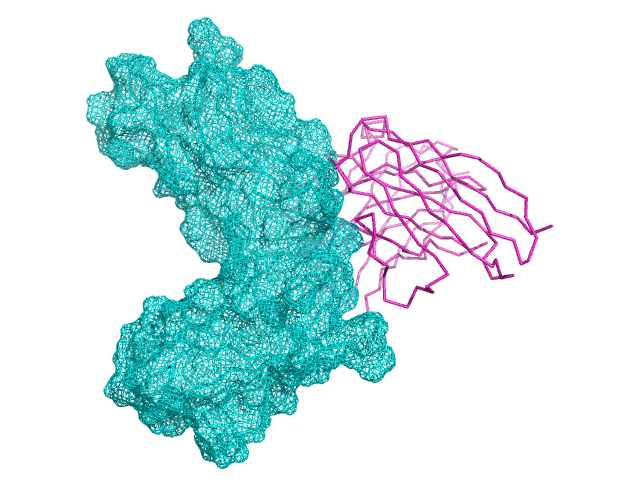
6. Final data
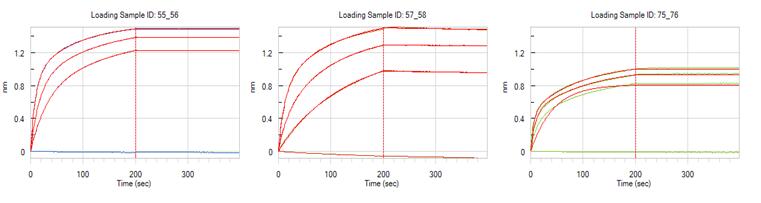
- Protein A based capture approach
- Anti-human Fc based capture approach
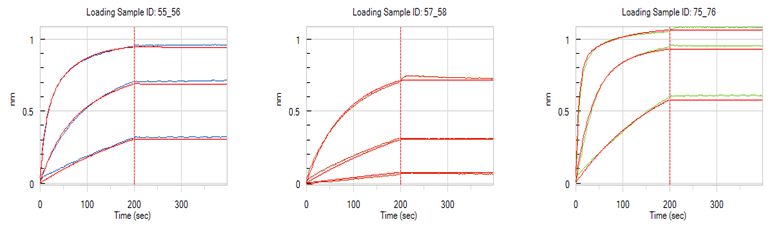
Designed 1~100 antibody sequences for the antigen, finding hits with affinity greater than 10-10M
Affinity test results
| Loading Sample ID | kon(1/Ms) | kdis(1/s) | KD (M) | Full R^2 |
|---|---|---|---|---|
| 55_56 | 9.28E+03 | 2.70E-07 | 2.91E-11 | 0.992429 |
| 57_58 | 3.76E+03 | 5.60E-07 | 1.49E-10 | 0.995068 |
| Positive control | 7.29E+04 | 8.90E-07 | 1.22E-11 | 0.996965 |

