Synbio Technologies can quickly and accurately sequence and analyze antibody sequences with our advanced Geno™ Ab Platform. Using this platform, we can perform high quality sequencing of multi-species’ (including mice, rats, Armenian hamsters, camels, rabbits, equines, bovines, canines, birds, etc.) monoclonal antibodies. The Syno® Ab Platform is a powerful computer-aided tool and has the capabilities to perform de novo sequencing analysis that may come from various antibody libraries, making this platform very applicable. Additionally, our experienced bioinformatics analysis team can also provide professional data mining and annotation.
Competitive Advantages
- We use custom primer sets and special reagents developed in-house to ensure high success rates.
- Our unique internal data mining tool can remove redundancies and extract information of target antibody sequence.
- Diversified NGS analysis platform has the powerful ability for antibody gene analysis, resulting in a high quality output.
- Complete bioinformatics analysis removes redundant sequence data, and deeply integrates the high-value information.
Service Specifications
| Service | Description | Turnaround Time | Deliverables | Price |
|---|---|---|---|---|
| Antibody Sequencing Bioinformatics Analysis | Sequencing data mining, Antibody information analysis, etc. | 7 – 10 Business Days | Analysis Reports | Quote |
Standard Antibody Sequencing Service Process
Case Study
Synbio Technologies’ immune repertoire sequencing and bioinformatics analysis:
- Library Construction
Synbio Technologies performed 5’ RACE reverse transcription of the extracted RNA, amplified, and constructed the library. - Library Quality Control
The Qubit 2.0 Fluorometer was used to perform preliminary quantification after the library construction. After dilution, insert size detection was performed on the library based on the Agilent 2100 bioanalyzer. Only when meeting expected requirements, the effective concentration of the library (>2nM) was accurately quantified by qPCR to ensure the quality of the library. - Library Sequencing
Different libraries were pooled and sequenced by Illumina Miseq/HiSeq according to the effective concentration and the required amount of target data. Four fluorescent labeled dNTPs, DNA polymerase, and adaptors were added to the flow cell for the amplification. When each sequence cluster extended and generated the complementary chain, the corresponding fluorescent signals were released and captured after each new dNTP was added. The optical signals were then transformed into sequencing peaks by professional software to obtain the sequence information of the fragments. - Data Analysis
Quality assessment of sequencing data.
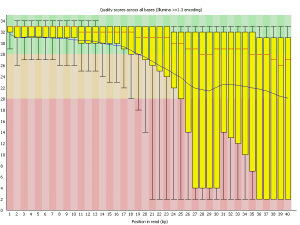
Per base sequence quality
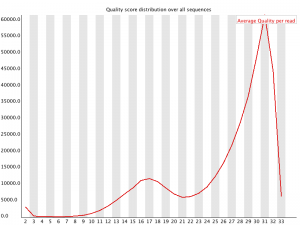
Per Sequence Quality Scores
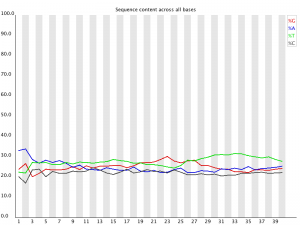
Per Base Sequence Content
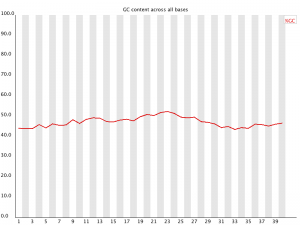
Per Base GC Content
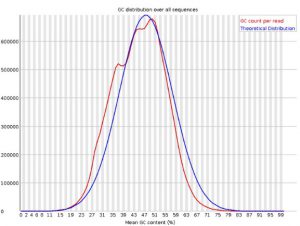
Per Sequence GC Content
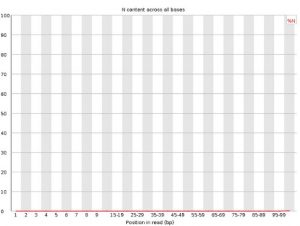
Per Base N Content
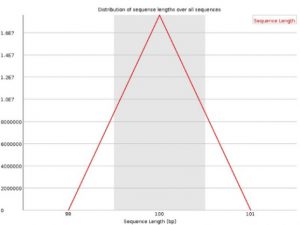
Sequence Length Distributionreads
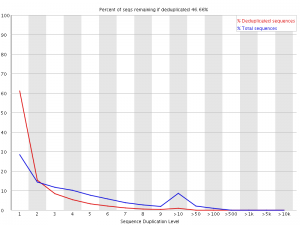
Duplicate Sequences

