Creating variant libraries is an important technique to study gene function, and it is widely used in protein characteristic improvement, protein directed evolution, antibody sequence optimization, affinity maturation, target discovery, etc. Researchers can change specific protein sites or DNA regulatory regions according to different purposes, forming a variant library containing tens of thousands of variant sequences while controlling the diversity of sequences and improving screening efficiency.
Synbio Technologies is committed to providing customers around the world with efficient and accurate precision variant library synthesis services, including scanning variant library, saturated variant library, custom variant library, and more. With more than ten years of experience in library synthesis of all complexities, we offer the industry’s most efficient and cost-effective solutions.
Genetic codons have degeneracy, that is, an amino acid can be encoded by more than one triplet codon. Using the degeneracy of genetic code can not only maintain the diversity of amino acid sequences, but also reduce the redundancy of codons and improve screening efficiency.
Competitive Advantages
• Quick & efficient product delivery to promote downstream screening
• The synthesis technology is stable and the quality control is strict
• 100% coverage of 20 amino acids fully ensures the diversity of the library
Case Study 1
The NNK library is a classic degenerate mutation library. The NNK degenerate primers used contain 32 (4x4x2) codon combinations (N= A/C/G/T, K= G/T), covering all 20 amino acids, and can saturate amino acids at any site.
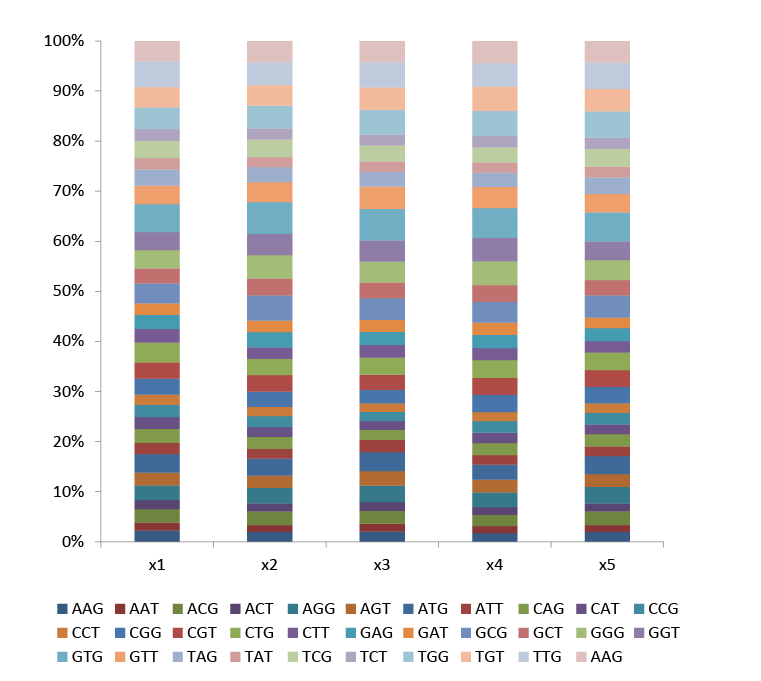
Fig.1 The proportion of each codon contained in the five NNK sites in the NNK library
Primers synthesized with a single nucleotide will inevitably produce redundant mutations and termination codons at the mutation site. The trimer phosphoramidites formed by connecting the three nucleotides in a fixed order can correspond to a single amino acid. It can not only introduce the mixture of all 20 amino acids at any position of the mutated sequence, but also avoids the problems of codon offset, frame shift mutation, and controlling the incorporation of termination codons.
Precise variant libraries designed with the trimer mixture as the element, also known as trimer libraries, can ensure that the amino acids at the mutation site are composed according to the designed ratio to meet the amino acid diversity and codon preference of the specific site. Trimer libraries are an efficient solution for complex libraries and can further improve the accuracy of library screening.
Competitive Advantages
• Cost-effective solutions
• Reduces difficulty of downstream screening
• High synthesis efficiency and accurate sequence control
Case Study 1
Trimer libraries can be used for site saturation mutation to distribute 20 amino acids in equal proportion, further reducing codon redundancy and removing termination codons in the library.
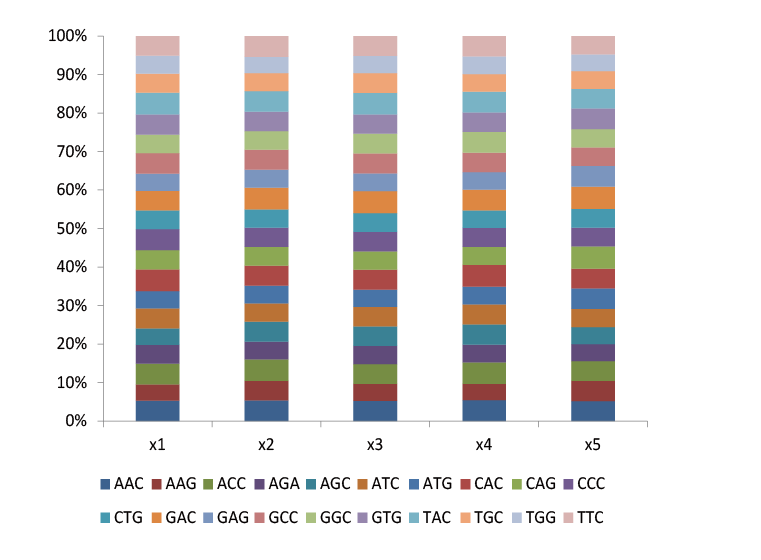
Fig.1 Trimer libraries are also a saturated mutation library with equal proportion distribution of each amino acid. The proportion of each codon contained in the five sites.
Case Study 2
Trimer libraries can also customize each site to be composed of different ratios of amino acids. Among the four Trimer sites, for example, the X1 Trimer site is A (80%) : G (3%) : R (3%) : S (3%): T (8%) : V (3%)
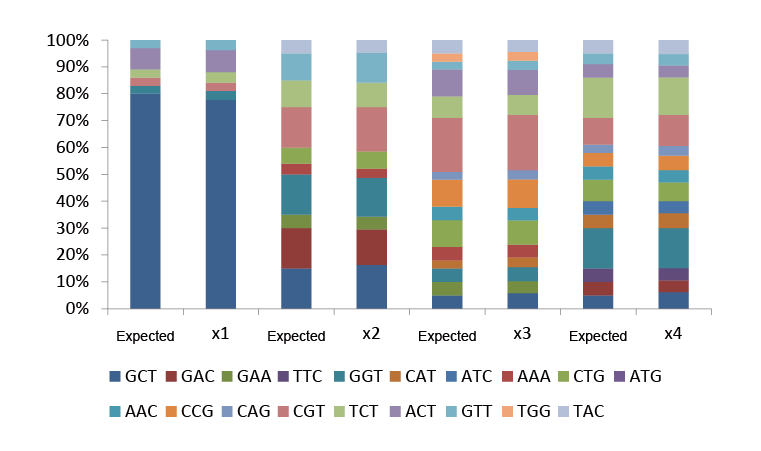
Fig.2 Types and proportions of custom amino acids
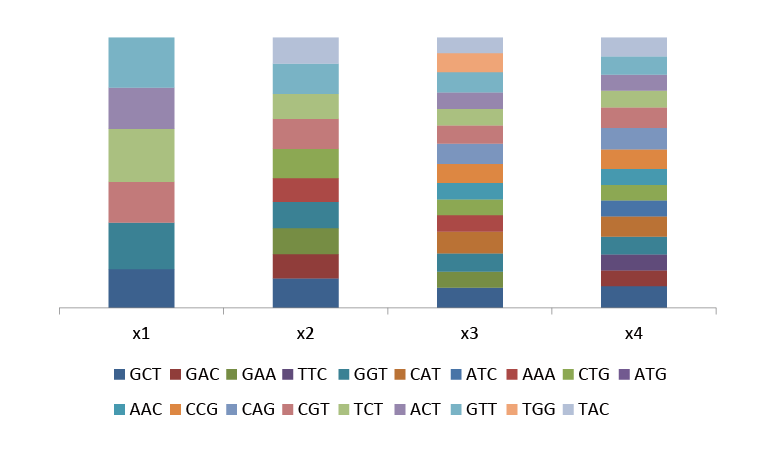
Fig.3 Ratio between actual and theoretical proportion of each amino acid

Accurate Sequence Control

Synthesis Efficiency

Affordable

Highly Customized

